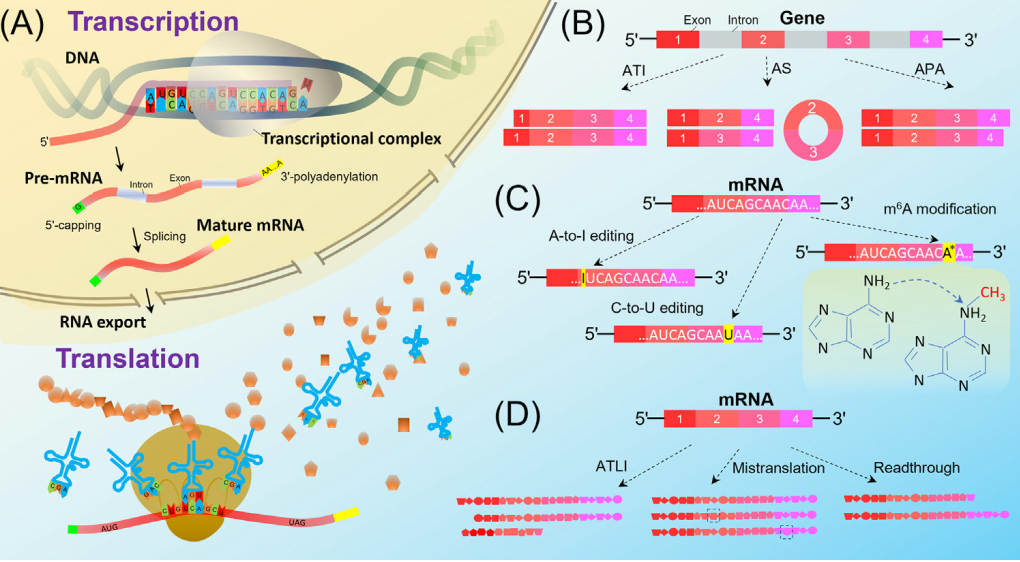
Genomic studies have shown that a single gene can give rise to multiple RNA molecules due to variable transcription initiation, splicing, and termination. Furthermore, enzymatic editing and modification can alter RNA bases or even its sequence, ultimately leading to changes in the translated proteins. During translation, variability in initiation and termination can also result in different protein products. Thus, a single gene can generate a wide array of RNA and protein molecules through transcription and translation. But, is this diversity in gene products a result of finely tuned gene regulation, or merely errors in gene expression? Some studies suggest that, in certain cases, gene product diversity may be beneficial, supporting the adaptive hypothesis favored by many researchers. However, through genome-wide analyses, we have found stronger support for the molecular error hypothesis, which posits that such diversity largely arises from errors during gene expression and is mostly nonfunctional or even deleterious.
Our lab focuses on the theoretical, mechanistic, and applied aspects of RNA and protein diversity (e.g., alternative splicing, back-splicing, alternative polyadenylation, alternative transcription starts, RNA modifications, RNA editing, alternative translation initiation, etc.). Using multi-omics data and an integrative approach that combines molecular and cellular biology, bioinformatics, and evolutionary genomics, we conduct research in the following three areas:
1) Theory and mechanism: refining the molecular error theory of RNA and protein diversity and uncovering its genomic patterns and evolutionary mechanisms.
2) Diseases: investigating how disruptions in RNA and protein diversity contribute to diseases (e.g., anxiety, depression, Alzheimer’s disease, and cancer).
3) Environmental adaptation: identifying atypical RNAs and proteins that play key roles in environmental adaptation, such as those found in pathogenic fungi.