ABOUT RNA Transcriptional Diversity
RNA transcriptional diversity represents a fundamental characteristic of dynamic genome expression, reflecting the complexity and adaptability of eukaryotic gene regulatory networks. Through precise control of transcription initiation, splicing processing, and termination, a single genetic locus can generate structurally diverse and functionally distinct RNA isoforms. This multi-dimensional regulatory mechanism not only dramatically expands the coding potential of limited genomes - enabling organisms to meet complex physiological demands with relatively compact genetic information - but also provides a flexible regulatory framework for critical biological processes including cellular differentiation, environmental adaptation, and stress responses.
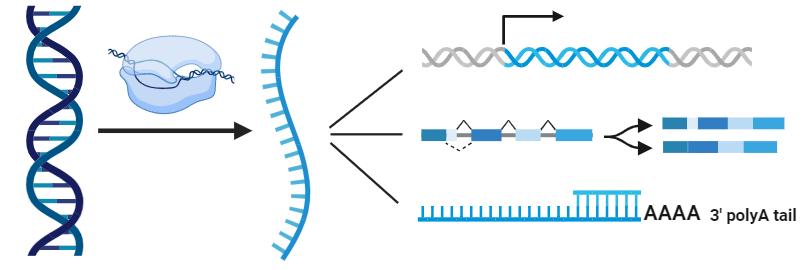
About ATS
Alternative Transcription Start Sites (ATS) mean that during gene transcription, RNA polymerase can start transcription from different locations of gene transcription start site (TSS). This results in the phenomenon of transcripts with different 5' end structures and functions. Different transcripts produced by ATS can translate protein isomers with different structures and functions, which greatly increases the complexity of proteome.
About AS
RNA Alternative Splicing (AS) is an important mechanism in eukaryotic gene expression regulation, which means that different exons in pre-mRNA can be spliced in different combinations during the post-transcriptional processing of genes. Thus, many different mature mRNA transcripts are produced, which in turn translate different protein isomers. The main types are exon jumping, intron retention, and mutually exclusive exons.
About APA
RNA Alternative Polyadenylation (APA) is an important part of eukaryotic gene expression regulation. In eukaryotes, the precursor mRNA produced by gene transcription usually requires the addition of a poly-A tail at the 3' end, a process called polyadenylation. RNA polyadenylation diversity refers to the fact that poly-A tails can be added to transcripts of the same gene at different sites during the polyadenylation process, resulting in mature mRNA transcripts with different 3' end structures and lengths.